Difference between revisions of "Galaxy"
From Bioinformatics Core Wiki
| Line 6: | Line 6: | ||
= CRG local implementation = | = CRG local implementation = | ||
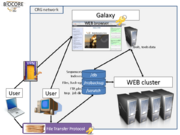
| − | + | Very soon will be available to all CRG users a local implementation of Galaxy including bioinformatics tools develloped in the CRG (Gem, T-Coffee and TrimAl for examples) as well as all tools provided by the central Galaxy. In order to protect data, CRG-Galaxy will only be available from the CRG network as describe in the Hardware schema. Data will be stored on the CRG file servers and all computations will run on a cluster of 72 computers. | |
| + | |||
| + | [[File:CRG-Galaxy-2.png|900px|snapshot of the CRG-Galaxy welcome page]] | ||
| − | |||
| − | |||
[[File:HardWare_Schema.png|thumb|Hardware schema of the CRG-Galaxy implementation]] | [[File:HardWare_Schema.png|thumb|Hardware schema of the CRG-Galaxy implementation]] | ||
Revision as of 11:50, 31 October 2012
Galaxy (http://wiki.g2.bx.psu.edu) is an open, web-based platform for accessible, reproducible, and transparent computational biomedical research.
- Accessible: Users without programming experience can easily specify parameters and run tools and workflows.
- Reproducible: Galaxy captures information so that any user can repeat and understand a complete computational analysis.
- Transparent: Users share and publish analyses via the web and create Pages, interactive, web-based documents that describe a complete analysis.
CRG local implementation
Very soon will be available to all CRG users a local implementation of Galaxy including bioinformatics tools develloped in the CRG (Gem, T-Coffee and TrimAl for examples) as well as all tools provided by the central Galaxy. In order to protect data, CRG-Galaxy will only be available from the CRG network as describe in the Hardware schema. Data will be stored on the CRG file servers and all computations will run on a cluster of 72 computers.