Difference between revisions of "ProjectsBlocks"
From Bioinformatics Core Wiki
Jponomarenko (Talk | contribs) |
Jponomarenko (Talk | contribs) |
||
| (7 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{{ProjectBlock | {{ProjectBlock | ||
|Title=Tool for CRISPR sgRNA Pairs Design | |Title=Tool for CRISPR sgRNA Pairs Design | ||
| − | |Description=CRISPETa is a web-tool for designing optimal pairs of sgRNAs for deletion of desired genomic regions; for example, lncRNAs. These target regions can be supplied in the BED or UCSC format. CRISPETa can be run on any number of targets - from one to thousands. At present, designs can be performed for five species, human, mouse, zebrafish, fruitfly, and worm. For more information and try the tool, visit [http://crispeta.crg.eu the CRISPETa website]. For curious, [https://en.wiktionary.org/wiki/crispeta ''crispeta'' means ''popcorn'', in Catalan]. | + | |Description=CRISPETa is a web-tool for designing optimal pairs of sgRNAs for deletion of desired genomic regions; for example, lncRNAs. These target regions can be supplied in the BED or UCSC format. CRISPETa can be run on any number of targets - from one to thousands. At present, designs can be performed for five species, human, mouse, zebrafish, fruitfly, and worm. For more information and try the tool, visit [http://crispeta.crg.eu the CRISPETa website]. For curious, [https://en.wiktionary.org/wiki/crispeta ''crispeta'' means ''popcorn'', in Catalan]. The paper about CRISPETa was published in [http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1005341 ''PLoS Comp. Biol.,'' 2017]. |
|Color=rgba(203, 75, 70, 0.5) | |Color=rgba(203, 75, 70, 0.5) | ||
|Image=Crispeta image.jpg | |Image=Crispeta image.jpg | ||
| Line 7: | Line 7: | ||
{{ProjectBlock | {{ProjectBlock | ||
|Title="Saca La Lengua" | |Title="Saca La Lengua" | ||
| − | |Description=''Stick Out Your Tongue'', or ''Saca la Lengua'' in | + | |Description=''Stick Out Your Tongue'', or ''Saca la Lengua'' in Spanish, is a citizen’s science project aiming to study the mouth’s microbiome of a large population of healthy young Spaniards and the relationship of microbiome profiles with environmental characteristics and lifestyle. The Bioinformatics Core is involved in processing of the 16S rRNA sequencing data, obtaining microbiome abundance profiles for each individual, setting up a website for the data analysis competition, and teaching students and public about bioinformatics and microbiome analysis. For more information, visit [http://www.sacalalengua.org the Saca La Lengua public website]. |
|Color=rgba(37, 174, 183, 0.24) | |Color=rgba(37, 174, 183, 0.24) | ||
|Image=Saca logo.png | |Image=Saca logo.png | ||
| Line 13: | Line 13: | ||
{{ProjectBlock | {{ProjectBlock | ||
|Title=Whole Genome Analysis of an Opitz C Syndrome Affected Family | |Title=Whole Genome Analysis of an Opitz C Syndrome Affected Family | ||
| − | |Description=The C syndrome, also known as the Opitz trigonocephaly syndrome, is a malformation syndrome characterized by trigonocephaly, severe mental retardation, hypotonia, variable cardiac defects, redundant skin, and dysmorphic facial features. Although this syndrome was described in 1969 by [http://medicine.utah.edu/faculty/mddetail.php?facultyID=u0030518 Dr. John M. Opitz], the causative genes and the mode of inheritance are still unknown [http://www.ncbi.nlm.nih.gov/pubmed/26768331 by Urreizti ''et al''., 2016]. The Bioinformatics Core collaborates with the geneticists from the University of Barcelona, Drs. Daniel Grinberg and Susana Balcells, and the Opitz C Association on analysis of the whole genomes of a patient and her parents to detect the putative causative mutations. | + | |Description=The C syndrome, also known as the Opitz trigonocephaly syndrome, is a malformation syndrome characterized by trigonocephaly, severe mental retardation, hypotonia, variable cardiac defects, redundant skin, and dysmorphic facial features. Although this syndrome was described in 1969 by [http://medicine.utah.edu/faculty/mddetail.php?facultyID=u0030518 Dr. John M. Opitz], the causative genes and the mode of inheritance are still unknown [http://www.ncbi.nlm.nih.gov/pubmed/26768331 by Urreizti ''et al''., 2016]. The Bioinformatics Core collaborates with the geneticists from the University of Barcelona, Drs. Daniel Grinberg and Susana Balcells, and the Opitz C Association on analysis of the whole genomes of a patient and her parents to detect the putative causative mutations. The paper describing the results of sequencing and a ''de novo'' nonsense mutation in MAGEL2 in the affected patient was published in [http://www.nature.com/articles/srep44138 ''Scientific Reports,'' 2017.] |
|Color=rgba(230, 205, 151, 0.41) | |Color=rgba(230, 205, 151, 0.41) | ||
|Image=Opitz image.jpg | |Image=Opitz image.jpg | ||
| Line 37: | Line 37: | ||
{{ProjectBlock | {{ProjectBlock | ||
|Title=''De novo'' Genome Assembly of Different Fly Species | |Title=''De novo'' Genome Assembly of Different Fly Species | ||
| − | |Description=The Bioinformatics Core collaborates with | + | |Description=The Bioinformatics Core collaborates with Karl Wotton (Exeter University, UK) to assemble and annotate the genomes of three non-drosophilid diptera, ''Clogmia albipunctata'', ''Megaselia abdita'', and ''Episyrphus balteatus''. In collaboration with the group of Dr. Yogi Jaeger (CRG) we previously published the ''de novo'' transcriptomes of these flies in [http://bmcgenomics.biomedcentral.com/articles/10.1186/1471-2164-14-123 ''BMC Genomics'', 2013]. |
|Color=rgba(30, 80, 225, 0.26) | |Color=rgba(30, 80, 225, 0.26) | ||
|Image=Screen Shot 2016-05-17 at 12.59.34.png | |Image=Screen Shot 2016-05-17 at 12.59.34.png | ||
| Line 55: | Line 55: | ||
{{ProjectBlock | {{ProjectBlock | ||
|Title=SuperFly Database | |Title=SuperFly Database | ||
| − | |Description=SuperFly is a database for the comparative analysis of spatio-temporal gene expression patterns and regulation in dipteran species (flies, midges, and mosquitoes). It currently hosts data on three species, the vinegar fly ''Drosophila melanogaster'', the scuttle fly ''Megaselia abdita'', and the moth midge ''Clogmia albipunctata''. The Bionformatics Core developed the database and the website to collect and visualize data. For more information, visit [http://superfly.crg.eu/ SuperFly]. The database was published in [http://www.ncbi.nlm.nih.gov/pubmed/25404137 ''NAR'', | + | |Description=SuperFly is a database for the comparative analysis of spatio-temporal gene expression patterns and regulation in dipteran species (flies, midges, and mosquitoes). It currently hosts data on three species, the vinegar fly ''Drosophila melanogaster'', the scuttle fly ''Megaselia abdita'', and the moth midge ''Clogmia albipunctata''. The Bionformatics Core developed the database and the website to collect and visualize data. For more information, visit [http://superfly.crg.eu/ SuperFly]. The database was published in [http://www.ncbi.nlm.nih.gov/pubmed/25404137 ''NAR'', 2015]. |
|Color=rgba(179, 123, 146, 0.31) | |Color=rgba(179, 123, 146, 0.31) | ||
|Image=Superfly-logo-home.jpg | |Image=Superfly-logo-home.jpg | ||
| Line 61: | Line 61: | ||
{{ProjectBlock | {{ProjectBlock | ||
|Title=Plant Resistance Genes Database | |Title=Plant Resistance Genes Database | ||
| − | |Description=The Plant Resistance Genes database, [http://prgdb.crg.eu/wiki/Main_Page PRGdb], is a comprehensive resource on pathogen resistance genes (R-genes) in plants. Initiated in 2009, it currently provides data on 112 known and | + | |Description=The Plant Resistance Genes database, [http://prgdb.crg.eu/wiki/Main_Page PRGdb], is a comprehensive resource on pathogen resistance genes (R-genes) in plants. Initiated in 2009, it currently provides data on 112 known and 104,310 putative R-genes in 233 plant species and conferring resistance to 122 pathogens. The Bionformatics Core participated in the project by redesigning the database website using the Semantic MediaWiki technologies. The databased was published in [http://nar.oxfordjournals.org/content/41/D1/D1167.long ''NAR'', 2013]. |
|Color=rgba(0, 199, 75, 0.39) | |Color=rgba(0, 199, 75, 0.39) | ||
|Image=PRG db logo.png | |Image=PRG db logo.png | ||
| Line 67: | Line 67: | ||
{{ProjectBlock | {{ProjectBlock | ||
|Title=Iberian Lynx Genome | |Title=Iberian Lynx Genome | ||
| − | |Description=This project, aimed at deciphering the genome of the Iberian lynx, is a collaborative effort of six research centers in Spain. It is part of the Project Zero program FGCSIC 2010 in Endangered Species and funded by Banco Santander and Fundación General CSIC. The Bioinformatics Core | + | |Description=This project, aimed at deciphering the genome of the Iberian lynx, is a collaborative effort of six research centers in Spain. It is part of the Project Zero program FGCSIC 2010 in Endangered Species and funded by Banco Santander and Fundación General CSIC. The Bioinformatics Core was involved in generation of a first annotated draft of the genome. For more information, visit [http://www.lynxgenomics.eu the project website]. The results were published in [http://genomebiology.biomedcentral.com/articles/10.1186/s13059-016-1090-1 ''Genome Biology'', 2016]. |
|Color=rgba(204, 190, 164, 0.73) | |Color=rgba(204, 190, 164, 0.73) | ||
|Image=Lynx.png | |Image=Lynx.png | ||
| Line 73: | Line 73: | ||
{{ProjectBlock | {{ProjectBlock | ||
|Title=Melon Genome | |Title=Melon Genome | ||
| − | |Description=Melon | + | |Description=Melon ''Cucumis melo L.'' is a cucurbit with high economic impact and a well-known and appreciated product in Europe. Spain is the fifth world producer and the main exporter of melon. To decipher the melon genome, the governmental foundation Genoma España funded the project ''MELONOMICS'', to which the Bioinformatics Core contributed by predicting small non-coding RNAs and pathogen resistant genes. For more information, visit [https://melonomics.net/ the MELONOMICS website]. The results were published in [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3406823/ ''PNAS'', 2012]. |
|Color=rgba(255, 149, 38, 0.32) | |Color=rgba(255, 149, 38, 0.32) | ||
|Image=Melonomics2.jpg | |Image=Melonomics2.jpg | ||
Latest revision as of 11:38, 14 March 2017
Tool for CRISPR sgRNA Pairs Design
CRISPETa is a web-tool for designing optimal pairs of sgRNAs for deletion of desired genomic regions; for example, lncRNAs. These target regions can be supplied in the BED or UCSC format. CRISPETa can be run on any number of targets - from one to thousands. At present, designs can be performed for five species, human, mouse, zebrafish, fruitfly, and worm. For more information and try the tool, visit the CRISPETa website. For curious, crispeta means popcorn, in Catalan. The paper about CRISPETa was published in PLoS Comp. Biol., 2017.
"Saca La Lengua"
Stick Out Your Tongue, or Saca la Lengua in Spanish, is a citizen’s science project aiming to study the mouth’s microbiome of a large population of healthy young Spaniards and the relationship of microbiome profiles with environmental characteristics and lifestyle. The Bioinformatics Core is involved in processing of the 16S rRNA sequencing data, obtaining microbiome abundance profiles for each individual, setting up a website for the data analysis competition, and teaching students and public about bioinformatics and microbiome analysis. For more information, visit the Saca La Lengua public website.
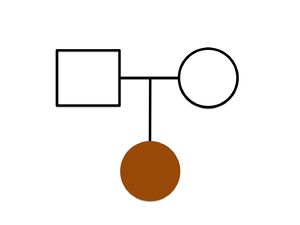
Whole Genome Analysis of an Opitz C Syndrome Affected Family
The C syndrome, also known as the Opitz trigonocephaly syndrome, is a malformation syndrome characterized by trigonocephaly, severe mental retardation, hypotonia, variable cardiac defects, redundant skin, and dysmorphic facial features. Although this syndrome was described in 1969 by Dr. John M. Opitz, the causative genes and the mode of inheritance are still unknown by Urreizti et al., 2016. The Bioinformatics Core collaborates with the geneticists from the University of Barcelona, Drs. Daniel Grinberg and Susana Balcells, and the Opitz C Association on analysis of the whole genomes of a patient and her parents to detect the putative causative mutations. The paper describing the results of sequencing and a de novo nonsense mutation in MAGEL2 in the affected patient was published in Scientific Reports, 2017.
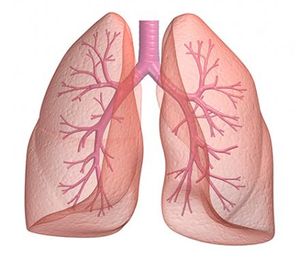
Lung Microbiomes of COPD Patients
CRG collaborates with the Hospital San Pau, Hospital Clinic, Hospital del Mar and other institutions in Barcelona on studies of the lung microbiomes of patients affected by chronic obstructive pulmonary disease (COPD). The Bioinformatics Core contributes to the project by processing and analyzing sequencing data. For more information, visit the Barcelona Respiratory Network website.
AnnoWiki
AnnoWiki is a Semantic MediaWiki based platform for hosting, managing and visualizing genome annotation data in a collaborative manner. This advanced Content Management System (CMS) is currently under development in the Bioinformatics Core. For more information, see our poster illustrating the application.
Wiki-based CMS for Core Facilities
The Bioinformatics Core developed several Semantic MediaWiki LIMSs (Laboratory Information Management System) to manage the laboratory's operations, data flow, and communication with users and external collaborators. One of such LIMS, ProteoWiki, is used by the CRG Proteomics Core.
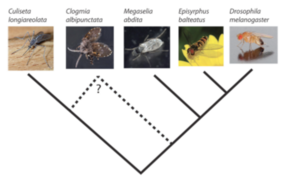
De novo Genome Assembly of Different Fly Species
The Bioinformatics Core collaborates with Karl Wotton (Exeter University, UK) to assemble and annotate the genomes of three non-drosophilid diptera, Clogmia albipunctata, Megaselia abdita, and Episyrphus balteatus. In collaboration with the group of Dr. Yogi Jaeger (CRG) we previously published the de novo transcriptomes of these flies in BMC Genomics, 2013.
Mycoplasma Database
MyMpn is a public resource for studying the human pathogen Mycoplasma pneumonia, a minimal bacterium causing lower respiratory tract infections. M. pneumonia is also used as a model unicellular organism for systems biology studies. The Bioinformatics Core developed MyMpn to host and analyze omics-data generated by the Design of Biological Systems Group (CRG) and collaborators at the EMBL and other institutions. For more information, visit the MyMpn website. The database was published in NAR, 2015.
Common Bean Genome
The Bioinformatics Core collaborated with an international consortium to decode the genome of the Mesoamerican common bean Phaseolus vulgarism. The core contributed to the project by identifying small ncRNAs and annotating genes predicted from the genome sequences. The findings were reported in Genome Biology, 2016.
SuperFly Database
SuperFly is a database for the comparative analysis of spatio-temporal gene expression patterns and regulation in dipteran species (flies, midges, and mosquitoes). It currently hosts data on three species, the vinegar fly Drosophila melanogaster, the scuttle fly Megaselia abdita, and the moth midge Clogmia albipunctata. The Bionformatics Core developed the database and the website to collect and visualize data. For more information, visit SuperFly. The database was published in NAR, 2015.
Plant Resistance Genes Database
The Plant Resistance Genes database, PRGdb, is a comprehensive resource on pathogen resistance genes (R-genes) in plants. Initiated in 2009, it currently provides data on 112 known and 104,310 putative R-genes in 233 plant species and conferring resistance to 122 pathogens. The Bionformatics Core participated in the project by redesigning the database website using the Semantic MediaWiki technologies. The databased was published in NAR, 2013.
Iberian Lynx Genome
This project, aimed at deciphering the genome of the Iberian lynx, is a collaborative effort of six research centers in Spain. It is part of the Project Zero program FGCSIC 2010 in Endangered Species and funded by Banco Santander and Fundación General CSIC. The Bioinformatics Core was involved in generation of a first annotated draft of the genome. For more information, visit the project website. The results were published in Genome Biology, 2016.
Melon Genome
Melon Cucumis melo L. is a cucurbit with high economic impact and a well-known and appreciated product in Europe. Spain is the fifth world producer and the main exporter of melon. To decipher the melon genome, the governmental foundation Genoma España funded the project MELONOMICS, to which the Bioinformatics Core contributed by predicting small non-coding RNAs and pathogen resistant genes. For more information, visit the MELONOMICS website. The results were published in PNAS, 2012.